import numpy as np
import pandas as pd
from econml.dr import LinearDRLearner
from joblib import Parallel, delayed
from lightgbm import LGBMClassifier, LGBMRegressor
from sklearn.linear_model import LinearRegression, LogisticRegression
import empirical_calibration as ec
import scipy
from typing import ListJeff Wooldridge recently posted about different estimators for treatment effects under unconfoundedness, and in particular emphasized a somewhat overlooked Regression Adjustment with inverse propensity weights method, which can be viewed as a (simple) instantiation of the Targeted Maximum Likelihood (TMLE) approach to estimation under unconfoundedness.
Exposition from Imbens (2004)
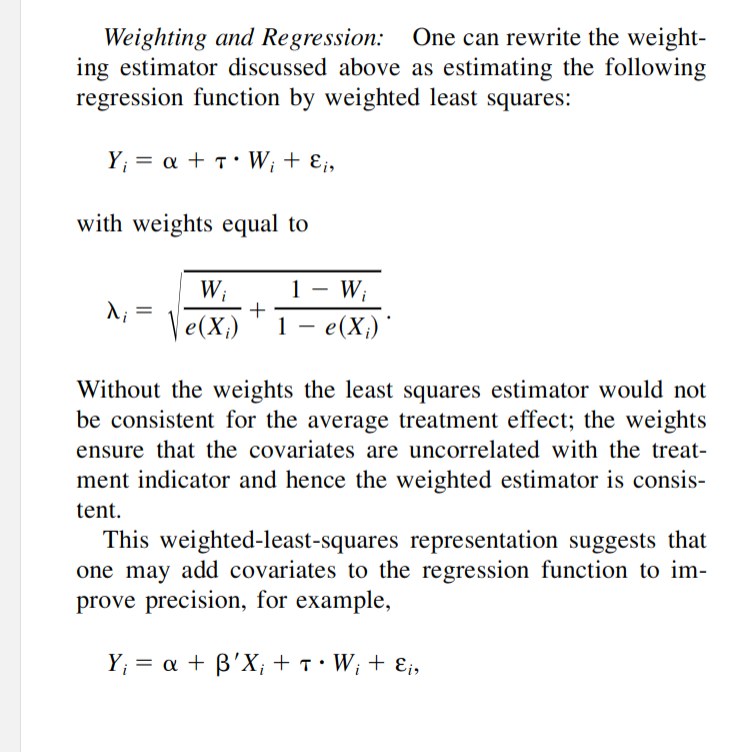
Here, we implement and benchmark some of these estimators and study their behaviour under different forms of mis-specification, and conclude that IPWRA generally works pretty well. It is effectively quite similar to entropy balancing weights in terms of bias and variance, which are also doubly robust in that they implicitly fit a linear model and logistic propensity score.
utils
DGP to generate a dataframe generated from a user-supplied pscore, outcome, and heterogeneous effects function.
# %% DGP
def binary_dgp(n, k, pscore_fn, tau_fn, outcome_fn, k_cat=1):
Sigma = np.random.uniform(-1, 1, (k, k))
Sigma = Sigma @ Sigma.T
Xnum = np.random.multivariate_normal(np.zeros(k), Sigma, n)
# generate categorical variables
Xcat = np.random.binomial(1, 0.5, (n, k_cat))
X = np.c_[Xnum, Xcat]
W = np.random.binomial(1, pscore_fn(X), n).astype(int)
Y = outcome_fn(X, W, tau_fn)
df = pd.DataFrame(
np.c_[Y, W, X], columns=["Y", "W"] + [f"X{i}" for i in range(k + 1)]
)
return dfdef sim_summariser(dat, truth=1):
return {
"bias": np.mean(dat, axis=0) - truth,
"var": np.var(dat, axis=0),
"rmse": np.sqrt(np.mean((dat - truth) ** 2, axis=0)),
}Regression Adjustment + IPW
Omnibus function to compute Regression Adjustment, IPW, and IPW-RA. This is now available in aipyw.
from sklearn.preprocessing import StandardScaler
def ipwra(
y: np.ndarray,
w: np.ndarray,
X: np.ndarray,
estimand: str = "ATE",
typ: str = "both",
):
# standardize X
sca = StandardScaler()
Xtilde = sca.fit_transform(X)
# weight specification
if typ in ["both", "ipw"]:
lr = LogisticRegression(
C=1e12
) # make inverse reg strength big for no regularisation
lr.fit(X, w)
p = lr.predict_proba(X) # returns (1-e(x), e(x))
if estimand == "ATE":
wt = w / p[:, 1] + (1 - w) / p[:, 0]
elif estimand == "ATT":
rho = np.mean(w)
wt = p[:, 1] / rho + (1 - w) / p[:, 0]
# covariate matrix specification
if typ in ["both", "ra"]:
XX = np.c_[
np.ones(len(w)), w, Xtilde, w[:, np.newaxis] * Xtilde
] # design matrix has [1, W, X, W * X] where X is centered
else:
XX = np.c_[np.ones(len(w)), w] # pure IPW
if typ == "ra":
wt = None
return LinearRegression().fit(XX, y, sample_weight=wt).coef_[1] # w coefedit: an earlier version manually applied np.sqrt to the weights, which LinearRegression does internally. Τhis was incorrect and has been fixed.
User-facing instantiation that accepts a dataframe, and computes bootstrap confidence intervals by default.
def bootstrap_ipwra(
df: pd.DataFrame,
outcome: str = "Y",
treatment: str = "W",
covariates: List = [],
typ: str = "both",
alpha: float = 0.05,
n_iterations: int = 100,
n_jobs: int = -1,
):
if len(covariates) == 0: # use all remaining columns as covariates
covariates = df.columns.difference([outcome, treatment]).tolist()
point_estimate = ipwra(df[outcome].values, df[treatment].values, df[covariates].values, typ=typ)
if n_iterations == 0:
return {"est": point_estimate.round(4)}
def onerep():
bsamp = df.sample(n=len(df), replace=True)
return ipwra(bsamp[outcome].values, bsamp[treatment].values, bsamp[covariates].values, typ=typ)
# multi-thread bootstrap
replicates = Parallel(n_jobs=n_jobs)(delayed(onerep)() for _ in range(n_iterations))
replicates = np.array(replicates)
lb, ub = np.percentile(replicates, [alpha / 2 * 100, (1 - alpha / 2) * 100])
return {
"est": point_estimate.round(4),
f"{(1-alpha)*100}% CI (Bootstrap)": np.r_[lb, ub].round(4),
}Example of use:
def outcome_fn(x, w, taufn):
base_term = x[:, 0] + 2 * x[:, 1] + x[:, 3]
return taufn(x) * w + base_term + np.random.normal(0, 1, len(w))
def pscore_fn(x):
lin = x[:, 0] - x[:, 1] - x[:, 2] + x[:, 3] + np.random.normal(0, 1, x.shape[0])
return scipy.special.expit(lin)
df = binary_dgp(n = 10_000, k = 4, tau_fn = lambda x: 1,
pscore_fn = pscore_fn, outcome_fn = outcome_fn)
df.head()| Y | W | X0 | X1 | X2 | X3 | X4 | |
|---|---|---|---|---|---|---|---|
| 0 | 0.853669 | 1.0 | 0.540357 | -0.246628 | -1.208470 | -2.053303 | 1.0 |
| 1 | -2.828752 | 0.0 | 0.055509 | -0.042991 | -0.779749 | -1.748115 | 1.0 |
| 2 | -2.055299 | 1.0 | 0.288326 | -1.199592 | -0.606765 | -1.226635 | 1.0 |
| 3 | 3.797470 | 0.0 | -0.708268 | 1.232548 | 1.141870 | 1.584794 | 0.0 |
| 4 | 1.116264 | 1.0 | 0.317727 | -0.945817 | -0.163137 | 1.769615 | 1.0 |
# default args conform to simulated data; else supply variable names
bootstrap_ipwra(df){'est': 0.9953, '95.0% CI (Bootstrap)': array([0.9357, 1.0547])}Simulations
Our simulation machine accepts DGP parameters and returns a list of estimates. For the first 3 estimators, we use linear + logistic specifications that accommodate linear and logistic propensity scores, so any non-linearity generates misspecification. The last AIPW estimator uses boosting regression and classification and should (at slow rates) learn all functions, so will never be misspecified.
def onesim(**kwargs):
df = binary_dgp(**kwargs)
outcome_column, treatment_column = "Y", "W"
feature_columns = df.columns.difference([outcome_column, treatment_column]).tolist()
# naive
naive = df.groupby("W")["Y"].mean().diff()[1] # treat - ctrl
# ipw and ipwra
ra, ipw, ipwra = (
bootstrap_ipwra(df, n_iterations=0, typ="ra"),
bootstrap_ipwra(df, n_iterations=0, typ="ipw"),
bootstrap_ipwra(df, n_iterations=0, typ="both"),
)
# aipw
m = LinearDRLearner(
model_regression=LGBMRegressor(n_estimators=50, verbose=-1),
model_propensity=LGBMClassifier(n_estimators=50, verbose=-1),
)
m.fit(
Y=df["Y"],
T=df["W"],
W=df[feature_columns],
)
dml_est = m.ate_inference().mean_point
# balancing weights
qweights, _ = ec.maybe_exact_calibrate(
covariates=df.query("W==0")[feature_columns].values,
target_covariates=df.query("W==1")[feature_columns].values,
objective=ec.Objective.QUADRATIC,
)
eweights, _ = ec.maybe_exact_calibrate(
covariates=df.query("W==0")[feature_columns].values,
target_covariates=df.query("W==1")[feature_columns].values,
objective=ec.Objective.ENTROPY,
)
qb_est = df.query("W==1")["Y"].mean() - np.average(df.query("W==0")["Y"], weights=qweights)
eb_est = df.query("W==1")["Y"].mean() - np.average(df.query("W==0")["Y"], weights=eweights)
return np.r_[naive, ra["est"], ipw["est"], ipwra["est"], dml_est, qb_est, eb_est]tau_fn = lambda x: 1
n, k, nsim = 10_000, 3, 100
estimators = ['naive', 'ra', 'ipw', 'ipwra', 'aipw', 'qb', 'eb']both correctly specified
Linear outcome and logistic pscore.
def outcome_fn(x, w, taufn):
base_term = x[:, 0] + 2 * x[:, 1] + x[:, 3]
return taufn(x) * w + base_term + np.random.normal(0, 1, len(w))
def pscore_fn(x):
lin = x[:, 0] - x[:, 1] - x[:, 2] + x[:, 3] + np.random.normal(0, 1, x.shape[0])
return scipy.special.expit(lin)
onesim(n=n, k=k, pscore_fn=pscore_fn, tau_fn=tau_fn, outcome_fn=outcome_fn)array([-0.37865908, 1.0071 , 0.9765 , 0.9991 , 1.02255154,
0.9882471 , 0.98856997])linear_sims = np.array(
Parallel(n_jobs=-1)(
delayed(onesim)(
n=n, k=k, pscore_fn=pscore_fn, tau_fn=tau_fn, outcome_fn=outcome_fn
)
for _ in range(nsim)
)
)
linear_summary = sim_summariser(linear_sims)
pd.DataFrame.from_dict(linear_summary).set_axis(estimators)| bias | var | rmse | |
|---|---|---|---|
| naive | -0.333937 | 1.189322 | 1.140542 |
| ra | -0.000373 | 0.000628 | 0.025067 |
| ipw | -0.006068 | 0.005668 | 0.075530 |
| ipwra | -0.003223 | 0.000829 | 0.028967 |
| aipw | 0.011722 | 0.001381 | 0.038965 |
| qb | -0.003911 | 0.000931 | 0.030758 |
| eb | -0.005386 | 0.001297 | 0.036418 |
good weights don’t add much variance to IPWRA (over RA). IPW is quite bad even though the model is correctly specified.
correct outcome, incorrect propensity
introduce nonlinearity into the pscore.
def outcome_fn(x, w, taufn):
base_term = x[:, 0] + 2 * x[:, 1] + x[:, 3]
return taufn(x) * w + base_term + np.random.normal(0, 1, len(w))
def pscore_fn(x):
lin = (
-x[:, 0]
- x[:, 1]
- x[:, 2]
+ x[:, 3]
+ np.exp(x[:, 3])
+ x[:, 2] ** 2
+ 0.1 * x[:, 1] ** 3
+ x[:, 0] * x[:, 1]
+ np.random.normal(0, 1, x.shape[0])
)
return scipy.special.expit(lin)
nlinear_sims1 = np.array(
Parallel(n_jobs=12)(
delayed(onesim)(
n=n, k=k, pscore_fn=pscore_fn, tau_fn=tau_fn, outcome_fn=outcome_fn
)
for _ in range(nsim)
)
)
nlinear_summary1 = sim_summariser(nlinear_sims1)
pd.DataFrame.from_dict(nlinear_summary1).set_axis(estimators)| bias | var | rmse | |
|---|---|---|---|
| naive | -0.391805 | 0.367960 | 0.722130 |
| ra | 0.004664 | 0.001873 | 0.043533 |
| ipw | -0.375230 | 0.146017 | 0.535551 |
| ipwra | 0.000944 | 0.002958 | 0.054398 |
| aipw | 0.059299 | 0.035076 | 0.196450 |
| qb | 0.002870 | 0.003020 | 0.055026 |
| eb | -0.001717 | 0.004221 | 0.064991 |
Bad pscore does not hurt IPWRA in terms of bias, but increases variance a bit.
incorrect outcome, correct propensity
introduce non-linearity into the outcome model.
def outcome_fn(x, w, taufn):
base_term = (
x[:, 0]
+ 2 * x[:, 1]
+ x[:, 3]
+ np.exp(x[:, 0])
+ 2 * (x[:, 1] > 0.5)
+ x[:, 1] ** 2
+ np.sqrt(np.abs(x[:, 2]))
+ x[:, 0] * x[:, 2]
)
return taufn(x) * w + base_term + np.random.normal(0, 1, len(w))
def pscore_fn(x):
lin = (
-x[:, 0]
- x[:, 1]
- x[:, 2]
+ x[:, 3]
+ np.random.normal(0, 1, x.shape[0])
)
return scipy.special.expit(lin)
nlinear_sims2 = np.array(
Parallel(n_jobs=12)(
delayed(onesim)(
n=n, k=k, pscore_fn=pscore_fn, tau_fn=tau_fn, outcome_fn=outcome_fn
)
for _ in range(nsim)
)
)
nlinear_summary2 = sim_summariser(nlinear_sims2)
pd.DataFrame.from_dict(nlinear_summary2).set_axis(estimators)| bias | var | rmse | |
|---|---|---|---|
| naive | -2.913566 | 4.837368 | 3.650512 |
| ra | 0.328193 | 0.246748 | 0.595365 |
| ipw | -0.065889 | 0.107900 | 0.335024 |
| ipwra | 0.050050 | 0.036513 | 0.197530 |
| aipw | -0.033077 | 0.073476 | 0.273075 |
| qb | 0.494620 | 0.102425 | 0.589129 |
| eb | 0.092074 | 0.030125 | 0.196477 |
RA has some bias. IPWRA has lower bias, which suggests that the weights help a bit.
both incorrect
both outcome and propensity have non-linearity.
def outcome_fn(x, w, taufn):
base_term = (
x[:, 0]
+ 2 * x[:, 1]
+ x[:, 3]
+ np.exp(x[:, 0])
+ 2 * (x[:, 1] > 0.5)
+ x[:, 1] ** 2
+ np.sqrt(np.abs(x[:, 2]))
+ x[:, 0] * x[:, 2]
)
return taufn(x) * w + base_term + np.random.normal(0, 1, len(w))
def pscore_fn(x):
lin = (
x[:, 0]
- x[:, 1]
- x[:, 2]
+ x[:, 3]
+ np.exp(x[:, 3])
+ x[:, 2] ** 2
+ 0.1 * x[:, 1] ** 3
+ x[:, 0] * x[:, 1]
+ np.random.normal(0, 1, x.shape[0])
)
return scipy.special.expit(lin)
nlinear_sims3 = np.array(
Parallel(n_jobs=12)(
delayed(onesim)(
n=n, k=k, pscore_fn=pscore_fn, tau_fn=tau_fn, outcome_fn=outcome_fn
)
for _ in range(nsim)
)
)
nlinear_summary3 = sim_summariser(nlinear_sims3)
pd.DataFrame.from_dict(nlinear_summary3).set_axis(estimators)| bias | var | rmse | |
|---|---|---|---|
| naive | 0.769820 | 2.343603 | 1.713542 |
| ra | 0.058784 | 3.701395 | 1.924799 |
| ipw | -4.453326 | 128.446402 | 12.176967 |
| ipwra | 0.441008 | 7.651208 | 2.801017 |
| aipw | 0.059865 | 0.040356 | 0.209618 |
| qb | -0.392092 | 5.627199 | 2.404357 |
| eb | -1.021150 | 10.837197 | 3.446729 |
All bets are off, only AIPW with boosting nuisances works well.