Using Fortran and C code with Python¶
J.R. Johansson (jrjohansson at gmail.com)
The latest version of this IPython notebook lecture is available at http://github.com/jrjohansson/scientific-python-lectures.
The other notebooks in this lecture series are indexed at http://jrjohansson.github.io.
%pylab inline
from IPython.display import Image
The advantage of Python is that it is flexible and easy to program. The time it takes to setup a new calulation is therefore short. But for certain types of calculations Python (and any other interpreted language) can be very slow. It is particularly iterations over large arrays that is difficult to do efficiently.
Such calculations may be implemented in a compiled language such as C or Fortran. In Python it is relatively easy to call out to libraries with compiled C or Fortran code. In this lecture we will look at how to do that.
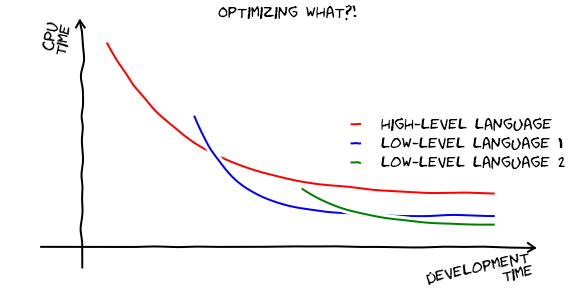
But before we go ahead and work on optimizing anything, it is always worthwhile to ask....
Image(filename='images/optimizing-what.png')
Fortran¶
F2PY¶
F2PY is a program that (almost) automatically wraps fortran code for use in Python: By using the f2py program we can compile fortran code into a module that we can import in a Python program.
F2PY is a part of NumPy, but you will also need to have a fortran compiler to run the examples below.
Example 0: scalar input, no output¶
%%file hellofortran.f
C File hellofortran.f
subroutine hellofortran (n)
integer n
do 100 i=0, n
print *, "Fortran says hello"
100 continue
end
Generate a python module using f2py:
!f2py -c -m hellofortran hellofortran.f
Example of a python script that use the module:
%%file hello.py
import hellofortran
hellofortran.hellofortran(5)
# run the script
!python hello.py
Example 1: vector input and scalar output¶
%%file dprod.f
subroutine dprod(x, y, n)
double precision x(n), y
y = 1.0
do 100 i=1, n
y = y * x(i)
100 continue
end
!rm -f dprod.pyf
!f2py -m dprod -h dprod.pyf dprod.f
The f2py program generated a module declaration file called dsum.pyf. Let's look what's in it:
!cat dprod.pyf
The module does not know what Fortran subroutine arguments is input and output, so we need to manually edit the module declaration files and mark output variables with intent(out) and input variable with intent(in):
%%file dprod.pyf
python module dprod ! in
interface ! in :dprod
subroutine dprod(x,y,n) ! in :dprod:dprod.f
double precision dimension(n), intent(in) :: x
double precision, intent(out) :: y
integer, optional,check(len(x)>=n),depend(x),intent(in) :: n=len(x)
end subroutine dprod
end interface
end python module dprod
Compile the fortran code into a module that can be included in python:
!f2py -c dprod.pyf dprod.f
Using the module from Python¶
import dprod
help(dprod)
dprod.dprod(arange(1,50))
# compare to numpy
prod(arange(1.0,50.0))
dprod.dprod(arange(1,10), 5) # only the 5 first elements
Compare performance:
xvec = rand(500)
timeit dprod.dprod(xvec)
timeit xvec.prod()
Example 2: cummulative sum, vector input and vector output¶
The cummulative sum function for an array of data is a good example of a loop intense algorithm: Loop through a vector and store the cummulative sum in another vector.
# simple python algorithm: example of a SLOW implementation
# Why? Because the loop is implemented in python.
def py_dcumsum(a):
b = empty_like(a)
b[0] = a[0]
for n in range(1,len(a)):
b[n] = b[n-1]+a[n]
return b
Fortran subroutine for the same thing: here we have added the intent(in) and intent(out) as comment lines in the original fortran code, so we do not need to manually edit the fortran module declaration file generated by f2py.
%%file dcumsum.f
c File dcumsum.f
subroutine dcumsum(a, b, n)
double precision a(n)
double precision b(n)
integer n
cf2py intent(in) :: a
cf2py intent(out) :: b
cf2py intent(hide) :: n
b(1) = a(1)
do 100 i=2, n
b(i) = b(i-1) + a(i)
100 continue
end
We can directly compile the fortran code to a python module:
!f2py -c dcumsum.f -m dcumsum
import dcumsum
a = array([1.0,2.0,3.0,4.0,5.0,6.0,7.0,8.0])
py_dcumsum(a)
dcumsum.dcumsum(a)
cumsum(a)
Benchmark the different implementations:
a = rand(10000)
timeit py_dcumsum(a)
timeit dcumsum.dcumsum(a)
timeit a.cumsum()
Further reading¶
C¶
ctypes¶
ctypes is a Python library for calling out to C code. It is not as automatic as f2py, and we manually need to load the library and set properties such as the functions return and argument types. On the otherhand we do not need to touch the C code at all.
%%file functions.c
#include <stdio.h>
void hello(int n);
double dprod(double *x, int n);
void dcumsum(double *a, double *b, int n);
void
hello(int n)
{
int i;
for (i = 0; i < n; i++)
{
printf("C says hello\n");
}
}
double
dprod(double *x, int n)
{
int i;
double y = 1.0;
for (i = 0; i < n; i++)
{
y *= x[i];
}
return y;
}
void
dcumsum(double *a, double *b, int n)
{
int i;
b[0] = a[0];
for (i = 1; i < n; i++)
{
b[i] = a[i] + b[i-1];
}
}
Compile the C file into a shared library:
!gcc -c -Wall -O2 -Wall -ansi -pedantic -fPIC -o functions.o functions.c
!gcc -o libfunctions.so -shared functions.o
The result is a compiled shared library libfunctions.so:
!file libfunctions.so
Now we need to write wrapper functions to access the C library: To load the library we use the ctypes package, which included in the Python standard library (with extensions from numpy for passing arrays to C). Then we manually set the types of the argument and return values (no automatic code inspection here!).
%%file functions.py
import numpy
import ctypes
_libfunctions = numpy.ctypeslib.load_library('libfunctions', '.')
_libfunctions.hello.argtypes = [ctypes.c_int]
_libfunctions.hello.restype = ctypes.c_void_p
_libfunctions.dprod.argtypes = [numpy.ctypeslib.ndpointer(dtype=numpy.float), ctypes.c_int]
_libfunctions.dprod.restype = ctypes.c_double
_libfunctions.dcumsum.argtypes = [numpy.ctypeslib.ndpointer(dtype=numpy.float), numpy.ctypeslib.ndpointer(dtype=numpy.float), ctypes.c_int]
_libfunctions.dcumsum.restype = ctypes.c_void_p
def hello(n):
return _libfunctions.hello(int(n))
def dprod(x, n=None):
if n is None:
n = len(x)
x = numpy.asarray(x, dtype=numpy.float)
return _libfunctions.dprod(x, int(n))
def dcumsum(a, n):
a = numpy.asarray(a, dtype=numpy.float)
b = numpy.empty(len(a), dtype=numpy.float)
_libfunctions.dcumsum(a, b, int(n))
return b
%%file run_hello_c.py
import functions
functions.hello(3)
!python run_hello_c.py
import functions
Product function:¶
functions.dprod([1,2,3,4,5])
Cummulative sum:¶
a = rand(100000)
res_c = functions.dcumsum(a, len(a))
res_fortran = dcumsum.dcumsum(a)
res_c - res_fortran
Simple benchmark¶
timeit functions.dcumsum(a, len(a))
timeit dcumsum.dcumsum(a)
timeit a.cumsum()
Further reading¶
Cython¶
A hybrid between python and C that can be compiled: Basically Python code with type declarations.
%%file cy_dcumsum.pyx
cimport numpy
def dcumsum(numpy.ndarray[numpy.float64_t, ndim=1] a, numpy.ndarray[numpy.float64_t, ndim=1] b):
cdef int i, n = len(a)
b[0] = a[0]
for i from 1 <= i < n:
b[i] = b[i-1] + a[i]
return b
A build file for generating C code and compiling it into a Python module.
%%file setup.py
from distutils.core import setup
from distutils.extension import Extension
from Cython.Distutils import build_ext
setup(
cmdclass = {'build_ext': build_ext},
ext_modules = [Extension("cy_dcumsum", ["cy_dcumsum.pyx"])]
)
!python setup.py build_ext --inplace
import cy_dcumsum
a = array([1,2,3,4], dtype=float)
b = empty_like(a)
cy_dcumsum.dcumsum(a,b)
b
a = array([1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0])
b = empty_like(a)
cy_dcumsum.dcumsum(a, b)
b
py_dcumsum(a)
a = rand(100000)
b = empty_like(a)
timeit py_dcumsum(a)
timeit cy_dcumsum.dcumsum(a,b)
Cython in the IPython notebook¶
When working with the IPython (especially in the notebook), there is a more convenient way of compiling and loading Cython code. Using the %%cython IPython magic (command to IPython), we can simply type the Cython code in a code cell and let IPython take care of the conversion to C code, compilation and loading of the function. To be able to use the %%cython magic, we first need to load the extension cythonmagic:
%load_ext cythonmagic
%%cython
cimport numpy
def cy_dcumsum2(numpy.ndarray[numpy.float64_t, ndim=1] a, numpy.ndarray[numpy.float64_t, ndim=1] b):
cdef int i, n = len(a)
b[0] = a[0]
for i from 1 <= i < n:
b[i] = b[i-1] + a[i]
return b
timeit cy_dcumsum2(a,b)
Further reading¶
Versions¶
%reload_ext version_information
%version_information ctypes, Cython