Databases in R¶
In [43]:
rm(list = ls())
library(LalRUtils)
libreq(data.table, magrittr, tidyverse, janitor, huxtable, knitr, tictoc)
theme_set(lal_plot_theme_d())
options(repr.plot.width = 15, repr.plot.height=12)
Basics¶
In [44]:
libreq(tidyverse, DBI, dbplyr, RSQLite, bigrquery, hrbrthemes, nycflights13, glue)
In [45]:
con <- dbConnect(RSQLite::SQLite(), path = ":memory:")
In [46]:
copy_to(
dest = con,
df = nycflights13::flights,
name = "flights",
temporary = FALSE,
indexes = list(
c("year", "month", "day"),
"carrier",
"tailnum",
"dest"
)
)
In [47]:
flights_db <- tbl(con, "flights")
flights_db %>% head
dplyr code that is SQL in disguise¶
In [48]:
tailnum_delay_db <-
flights_db %>%
group_by(tailnum) %>%
summarise(
mean_dep_delay = mean(dep_delay),
mean_arr_delay = mean(arr_delay),
n = n()
) %>%
arrange(desc(mean_arr_delay)) %>%
filter(n > 100)
tailnum_delay_db %>% show_query()
Run SQL code directly¶
Using glue to stitch together a query
In [50]:
## Some local R variables
tbl <- "flights"
d_var <- "dep_delay"
d_thresh <- 240
## The "glued" SQL query string
sql_query <-
glue_sql("
SELECT *
FROM {`tbl`}
WHERE ({`d_var`} > {d_thresh})
LIMIT 5
",
.con = con
)
## Run the query
dbGetQuery(con, sql_query)
In [51]:
dbDisconnect(con)
Manually setting up sqlite database from large CSV¶
Forked a nice function that handles the whole process by chunking the reading steps and writing to a sqlite database, which can then be processed using dplyr's lazy evaluation.
In [6]:
print(csv_to_sqlite)
In [7]:
help(csv_to_sqlite)
Example - All cabrides in NYC in 2014¶
In [8]:
libreq(gdata)
In [35]:
raw_path = "/home/alal/Downloads/Data_Drop/nyc_cabrides_2014.csv"
tmp = "/home/alal/tmp/db/"
sqlite_path= file.path(tmp, "NYC_cabs_2014.sqlite")
In [36]:
humanReadable(file.info(csv_path)$size)
examine slice of data
In [46]:
raw = fread(raw_path)
raw %>% glimpse
In [38]:
tic()
csv_to_sqlite(csv_path, sqlite_path, table_name = "cabs")
toc()
This took 3 hours and really was not worth it.
In [39]:
my_db <- src_sqlite(sqlite_path, create = FALSE)
cabs <- tbl(my_db, "cabs")
cabs %>% head()
In [42]:
cabs %>% group_by(pickup_date) %>% transmute(n_rides = n()) %>% collect() ->
n_rides
In [45]:
cabs %>% pull(pickup_date) %>% unique() %>% length()
In [7]:
libreq(bigrquery)
In [25]:
projid <- Sys.getenv("GCE_DEFAULT_PROJECT_ID")
bq_auth(email = "lal.apoorva@gmail.com",
path = "~/keys/sandbox.json")
Medicaid data¶
In [21]:
con <- dbConnect(
bigrquery::bigquery(),
project = "bigquery-public-data",
dataset = "medicare",
billing = projid
)
con
dbListTables(con)
In [22]:
ip_2011 = tbl(con, "inpatient_charges_2011")
In [24]:
ip_2011 %>% glimpse()
In [38]:
dbDisconnect(con)
All GCP samples¶
In [27]:
bq_con <- dbConnect(
bigrquery::bigquery(),
project = "publicdata",
dataset = "samples",
billing = projid
)
bq_con
dbListTables(bq_con)
Natality¶
In [29]:
natality <- tbl(bq_con, "natality")
natality %>% glimpse
In [30]:
bw <- natality %>%
group_by(year) %>%
summarise(weight_pounds = mean(weight_pounds, na.rm=T)) %>%
collect()
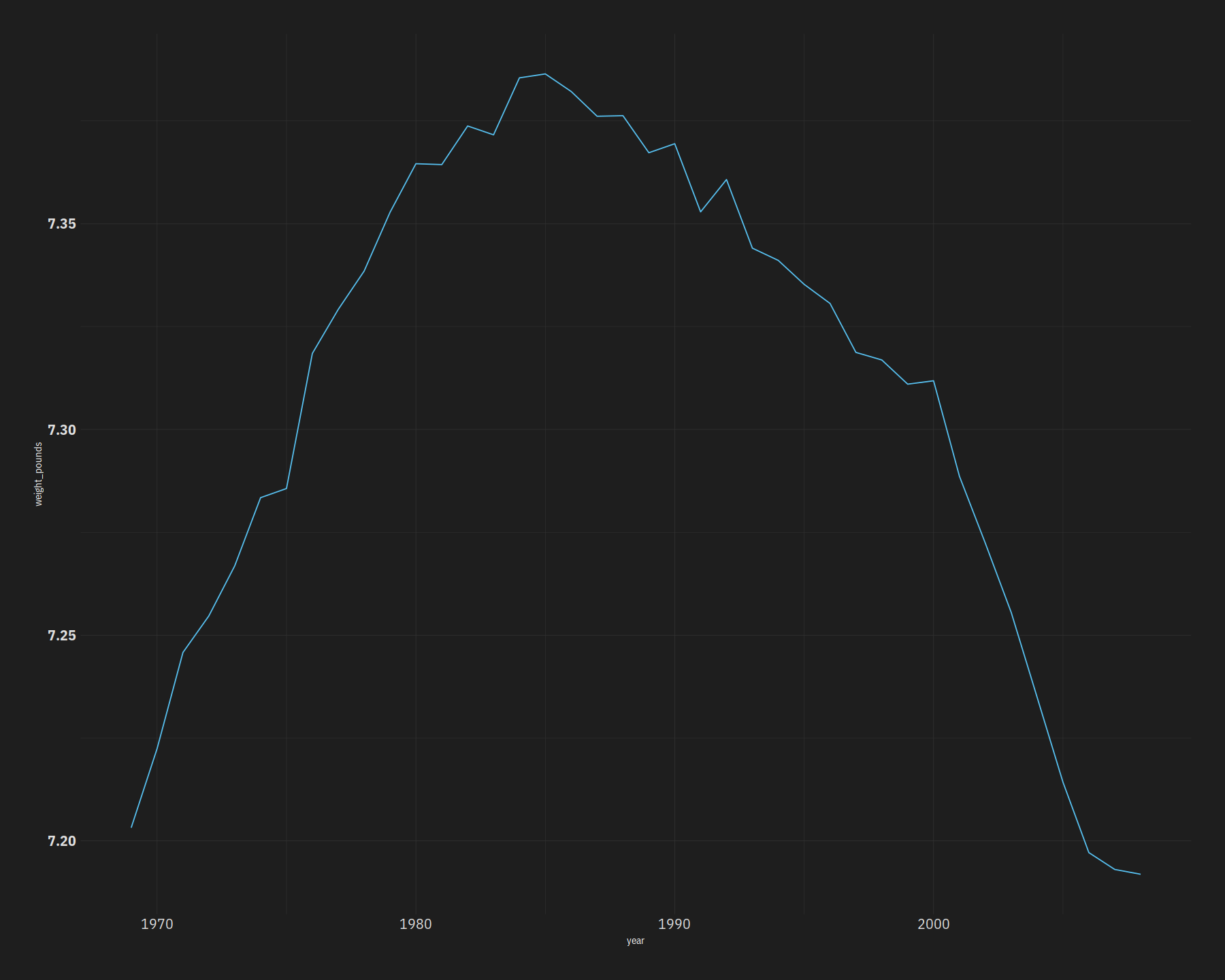
bw %>%
ggplot(aes(year, weight_pounds)) +
geom_line()
In [37]:
dbDisconnect(bq_con)
Fisheries¶
In [32]:
gfw_con <-
dbConnect(
bigrquery::bigquery(),
project = "global-fishing-watch",
dataset = "global_footprint_of_fisheries",
billing = projid
)
dbListTables(gfw_con)
In [33]:
effort <- tbl(gfw_con, "fishing_effort")
effort %>% glimpse
In [36]:
effort %>%
group_by(flag) %>%
summarise(total_fishing_hours = sum(fishing_hours, na.rm=T)) %>%
arrange(desc(total_fishing_hours)) %>%
collect() %>%
head(10)
In [39]:
## Define the desired bin resolution in degrees
resolution <- 0.5
effort %>%
filter(
`_PARTITIONTIME` >= "2016-01-01 00:00:00",
`_PARTITIONTIME` <= "2016-12-31 00:00:00"
) %>%
filter(fishing_hours > 0) %>%
mutate(
lat_bin = lat_bin/100,
lon_bin = lon_bin/100
) %>%
mutate(
lat_bin_center = floor(lat_bin/resolution)*resolution + 0.5*resolution,
lon_bin_center = floor(lon_bin/resolution)*resolution + 0.5*resolution
) %>%
group_by(lat_bin_center, lon_bin_center) %>%
summarise(fishing_hours = sum(fishing_hours, na.rm=T)) %>%
collect() ->
globe
In [41]:
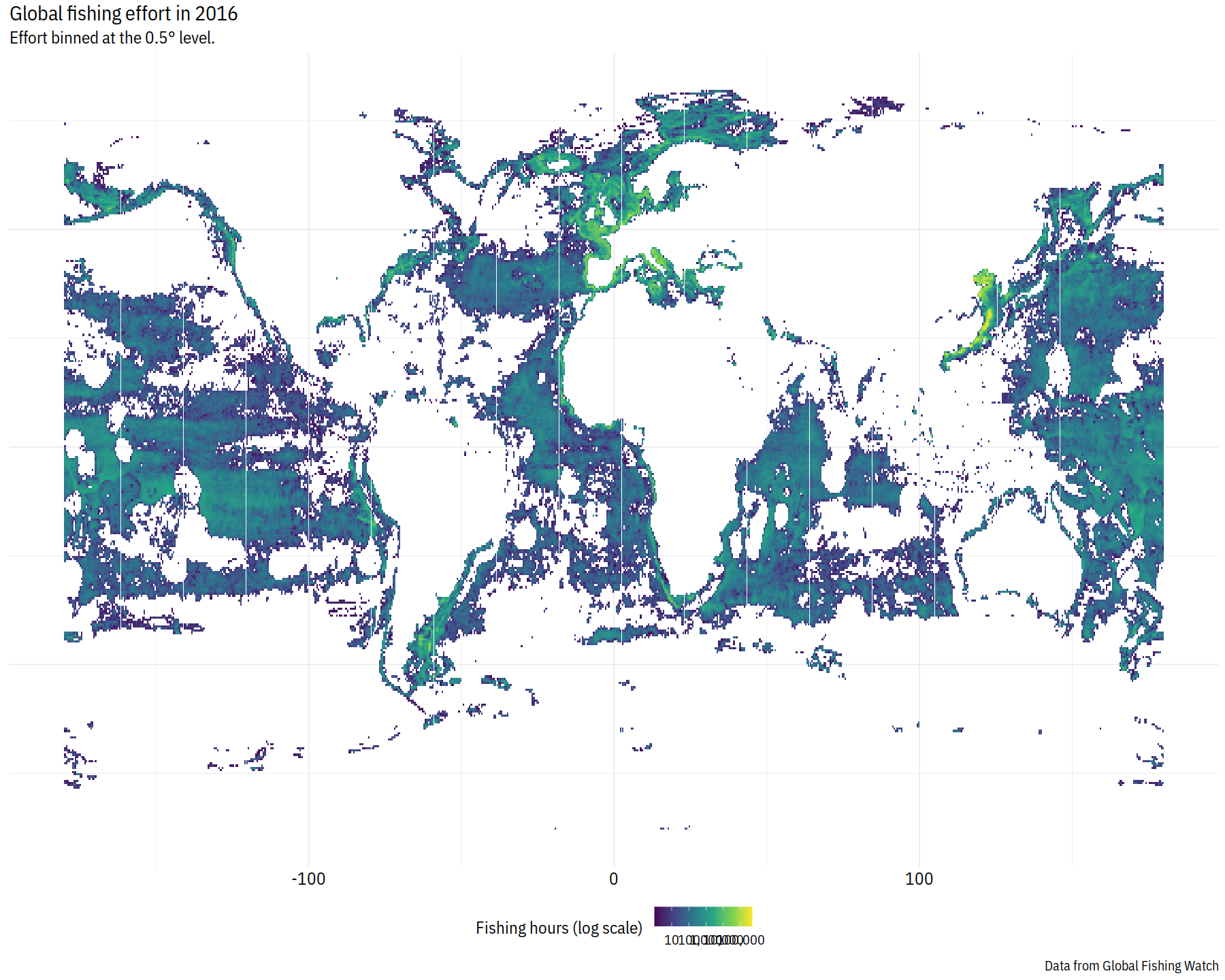
globe %>%
filter(fishing_hours > 1) %>%
ggplot() +
geom_tile(aes(x=lon_bin_center, y=lat_bin_center, fill=fishing_hours))+
scale_fill_viridis_c(
name = "Fishing hours (log scale)",
trans = "log",
breaks = scales::log_breaks(n = 5, base = 10),
labels = scales::comma
) +
labs(
title = "Global fishing effort in 2016",
subtitle = paste0("Effort binned at the ", resolution, "° level."),
y = NULL, x = NULL,
caption = "Data from Global Fishing Watch"
) +
lal_plot_theme() +
theme(axis.text=element_blank())
In [42]:
dbDisconnect(gfw_con)